BioSolveIT SeeSAR 13.1.0 | 132.7 mb
The Team BioSolveIT are proud to announce the next major version of SeeSAR 13.1.0 named ‘Midas’. This interactive tool allows drug designers to work with molecules interactively, for example, by docking ligands into the active sites of proteins to optimize hits.
SeeSAR ‘Midas’ version 13 what's new
SeeSAR ‘Midas’ version 13 was developed to be used as a powerful but user-friendly tool for the generation of goal-oriented results — with a single click! The novel ideation engine is called ‘MedChemesis’: a ligand mutation tool to create a series of close analogs to a query compound based on common medicinal chemistry reactions. MedChemesis can be accessed in the Inspirator Mode to sample the proximal chemical space around a compound for promising modifications (e.g., bioisosteric replacement of a carboxylic acid for a tetrazole group, introduction of a halogen or methyl group to increase potency, or replacement of a carbon for a nitrogen atom inside an aromatic ring system). With a total of 290 most common medicinal chemistry transformations, MedChemesis explores a plethora of analogs guided by binding site topology in the most efficient way — without the need of full enumeration of all possibilities. Alongside several quality-of-life software improvements, another great feature has been added to augment the on-screen design experience: Users can now visualize the target/protein surface. Several coloring and visualization options (based on lipophilicity, chain colors, opaque/transparent, …) are available to create meaningful figures based on your needs. The features does not end there: All 3D surface representations can be exported via SeeSAR glb file export to be used in presentations to visually transfer the core messages of your science!
What's New in SeeSAR 13 'Midas'?
Improvements to covalent docking
- Automatic detection of covalent warheads: SeeSAR now detects 36 most commonly used covalent warheads from input molecules and automatically transforms them to be used for covalent docking resulting in the creation of covalent protein-ligand complexes.
. The detection and transformation take place in the background once a covalent docking run has been initiated.
. Users can now avoid the tedious steps of having to manually edit molecules and adding linker atoms [R] at the appropriate location on input molecules.
. Molecules with linkers [R] are still supported for covalent docking as per usual.
- Automatic detection protein residues suitable for covalent docking:
. SeeSAR now detects the residues Cys, Ser, Thr, Tyr, Lys, Gln and Asn in the binding site and prepares them to be anchors for covalent attachment of molecules via docking, in the Docking Mode.
. Any residue (from the list above) suitable for covalent docking in the binding site is highlighted by a blue translucent capsule on the side-chain at which the covalent attachment could potentially happen.
. To select one of the residues for covalent docking, a single click on the corresponding capsule is sufficient. The assignment of the residue for covalent docking is visually indicated by the color of the capsule changing to pink.
. Note: SeeSAR will not determine the compatibility between the selected residue and the molecules in the docking library during the docking. All molecules from the docking library will be docked on the chosen residue.
Large-scale docking on external hardware: Introducing the external Docking Mode.
- The Docking mode switch can now be toggled to the right to find SeeSAR’s new mode for running large scale docking calculations external hardware such as a single powerful machine, or a cluster.
- The prerequisite for the successful usage of this mode is the configuration of HPSee, BioSolveIT’s high-performance computing framework, which is also introduced along with this release.
- Once HPSee has been installed on the external server (remote machines, clusters), access to the server can be configured via the newly added “Web Service” option within SeeSAR’s System Settings.
- The mode accepts a variety of inputs including molecules added from other modes, molecules loaded from external files, and also libraries uploaded to the external server via HPSee.
- All docking runs on this mode are run on the external hardware configured, and once the run has been initiated, users can switch to other modes in SeeSAR to carry out other tasks.
- HYDE optimization of the docked poses can also be done on the server if the user wishes by turning on the corresponding automatic calculation in the “Calculations” option in the System Settings.
- Note: Once a docking run has been initiated on this mode, it is recommended that the binding site of the protein used is not changed.
- Users are informed of the status of an ongoing docking run via a progress indicator on the play button.
- Users can cancel a run that has been started by clicking on the orange button which both terminates the run and deletes all results from the server.
- Users are informed about completion, of their initiated runs via appropriate notification messages delivered via the Message Center.
Usability improvements: Enhanced control on color and visibility.
- The Sequence View from the existing versions of SeeSAR is now more aptly named as the “Target View Control” panel. This section offers improved usability to control the visibility and color of the different components of the target macromolecules loaded into SeeSAR.
- The self-explanatory dropdown items “Visibility” and “Color” dictate which components on the target i.e., residues, chains, molecules, metals, and surfaces on which the intended color or visibility assignment is to take place.
Miscellaneous improvements
- When loading proteins into the Proteins or the Protein Editor mode, SeeSAR checks completeness of the structure and warns the user via an indicator on the protein entry on the table if the structure is incomplete
- A warning triangle on the PDB input field is shown if SeeSAR detects that the PDB info database is out of date.
- Automatic detection of covalent warheads: SeeSAR now detects 36 most commonly used covalent warheads from input molecules and automatically transforms them to be used for covalent docking resulting in the creation of covalent protein-ligand complexes.
. The detection and transformation take place in the background once a covalent docking run has been initiated.
. Users can now avoid the tedious steps of having to manually edit molecules and adding linker atoms [R] at the appropriate location on input molecules.
. Molecules with linkers [R] are still supported for covalent docking as per usual.
- Automatic detection protein residues suitable for covalent docking:
. SeeSAR now detects the residues Cys, Ser, Thr, Tyr, Lys, Gln and Asn in the binding site and prepares them to be anchors for covalent attachment of molecules via docking, in the Docking Mode.
. Any residue (from the list above) suitable for covalent docking in the binding site is highlighted by a blue translucent capsule on the side-chain at which the covalent attachment could potentially happen.
. To select one of the residues for covalent docking, a single click on the corresponding capsule is sufficient. The assignment of the residue for covalent docking is visually indicated by the color of the capsule changing to pink.
. Note: SeeSAR will not determine the compatibility between the selected residue and the molecules in the docking library during the docking. All molecules from the docking library will be docked on the chosen residue.
Large-scale docking on external hardware: Introducing the external Docking Mode.
- The Docking mode switch can now be toggled to the right to find SeeSAR’s new mode for running large scale docking calculations external hardware such as a single powerful machine, or a cluster.
- The prerequisite for the successful usage of this mode is the configuration of HPSee, BioSolveIT’s high-performance computing framework, which is also introduced along with this release.
- Once HPSee has been installed on the external server (remote machines, clusters), access to the server can be configured via the newly added “Web Service” option within SeeSAR’s System Settings.
- The mode accepts a variety of inputs including molecules added from other modes, molecules loaded from external files, and also libraries uploaded to the external server via HPSee.
- All docking runs on this mode are run on the external hardware configured, and once the run has been initiated, users can switch to other modes in SeeSAR to carry out other tasks.
- HYDE optimization of the docked poses can also be done on the server if the user wishes by turning on the corresponding automatic calculation in the “Calculations” option in the System Settings.
- Note: Once a docking run has been initiated on this mode, it is recommended that the binding site of the protein used is not changed.
- Users are informed of the status of an ongoing docking run via a progress indicator on the play button.
- Users can cancel a run that has been started by clicking on the orange button which both terminates the run and deletes all results from the server.
- Users are informed about completion, of their initiated runs via appropriate notification messages delivered via the Message Center.
Usability improvements: Enhanced control on color and visibility.
- The Sequence View from the existing versions of SeeSAR is now more aptly named as the “Target View Control” panel. This section offers improved usability to control the visibility and color of the different components of the target macromolecules loaded into SeeSAR.
- The self-explanatory dropdown items “Visibility” and “Color” dictate which components on the target i.e., residues, chains, molecules, metals, and surfaces on which the intended color or visibility assignment is to take place.
Miscellaneous improvements
- When loading proteins into the Proteins or the Protein Editor mode, SeeSAR checks completeness of the structure and warns the user via an indicator on the protein entry on the table if the structure is incomplete
- A warning triangle on the PDB input field is shown if SeeSAR detects that the PDB info database is out of date.
BioSolveIT SeeSAR allows drug designers to work with molecules interactively, for example, by docking ligands into the active sites of proteins to optimize hits. This interactive tool has been conceived to be easy to use, and will assist you in calculating physico-chemical properties, affinity estimations, and on-the-fly editing and hypothesis testing. You will learn how to optimize the tightness of fit and create meaningful graphics.
Introduction to SeeSAR: Drug Discovery with ‘Midas’
In this BioSolveIT workshop, we embark on a journey into the world of the drug discovery dashboard SeeSAR: A cutting-edge tool that has undergone a remarkable power gain over the years. During this workshop, we will offer a comprehensive introduction to its various operational “modes”. Modes, pleasingly designed, cover a wide array of tasks such as molecule editing, docking, and target-ligand complex assessment. Additionally to the structure-based methods, we will discuss how SeeSAR can facilitate ligand-based drug discovery (LBDD). The webinar represents a perfect opportunity for beginners to learn basic operations and gain insights into computer-aided drug discovery (CADD) with state-of-the-art software.
BioSolveIT is custom scientific software development company for virtual screening and lead discovery offers tools, services, and research collaborations. BioSolveIT provides world-renowned software products within the areas of ligand and structure-based drug design and is the pioneer of computational fragment-based ligand design. With a stellar scientific advisory board and founders from academia who intensely collaborate with pharma, BioSolveIT catalyzes products off of university research successes with proven pharmaceutical industry application.
Owner: BioSolveIT
Product Name: SeeSAR
Version: 13.1.0
Supported Architectures: x64
Website Home Page : www.biosolveit.de
Languages Supported: english
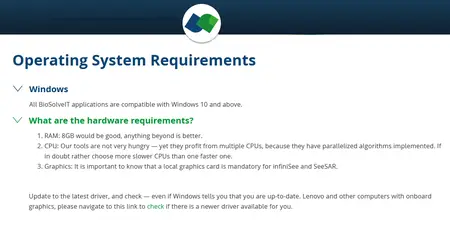
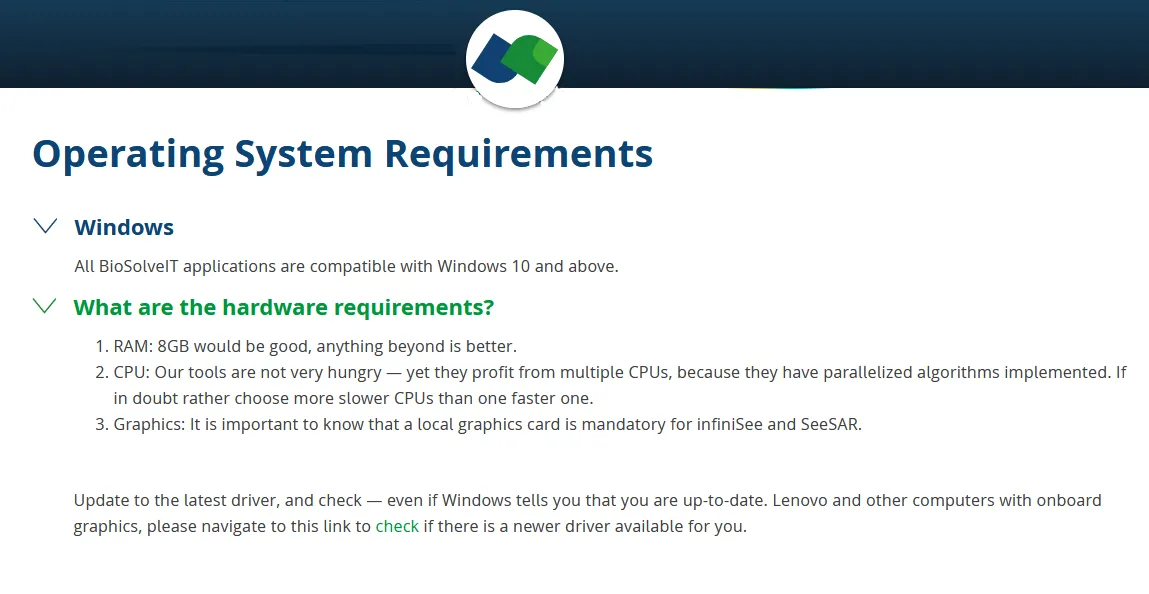
System Requirements: Windows *
Size: 132.7 mb
Please visit my blog
Added by 3% of the overall size of the archive of information for the restoration
No mirrors please
Added by 3% of the overall size of the archive of information for the restoration
No mirrors please