Master Metagenomics and Microbiome Data Analysis Using Linux
Published 7/2025
Duration: 3h 42m | .MP4 1280x720 30 fps(r) | AAC, 44100 Hz, 2ch | 1.75 GB
Genre: eLearning | Language: English
Published 7/2025
Duration: 3h 42m | .MP4 1280x720 30 fps(r) | AAC, 44100 Hz, 2ch | 1.75 GB
Genre: eLearning | Language: English
Learn to Analyze metagenomic and microbiome datasets using QIIME2, Kraken2, Bracken, and Linux-based pipelines
What you'll learn
- Understand the fundamentals of metagenomics and microbiome research, including 16S rRNA and shotgun sequencing approaches.
- Set up a complete Linux-based bioinformatics environment using WSL or VirtualBox for microbiome data analysis.
- Perform quality control and preprocessing of raw sequencing reads using command-line tools.
- Analyze 16S rRNA gene sequencing data using QIIME2 for diversity analysis and taxonomic profiling.
- Compute and visualize alpha and beta diversity metrics to assess microbial community structure.
- Perform taxonomic classification and differential abundance testing using QIIME2 and ANCOM.
- Use Kraken2 and Bracken to classify shotgun metagenomic reads and estimate microbial abundance.
- Visualize microbial composition interactively using Krona charts and QIIME2 visualizations.
- Interpret biological significance from taxonomic and diversity analysis results in real-world datasets.
- Build reproducible pipelines for end-to-end microbiome data analysis suitable for research or publication.
Requirements
- No prior experience in bioinformatics is required; this course is designed to be beginner-friendly and walks you through every step.
- Basic understanding of biology or microbiology is helpful, but not mandatory.
- A computer with at least 8GB of RAM (Windows, macOS, or Linux) to run tools smoothly.
- Willingness to use the Linux command line (we’ll guide you through it step-by-step).
- A curious mind and eagerness to explore the microbial world!
- Stable internet connection for downloading datasets and installing tools.
Description
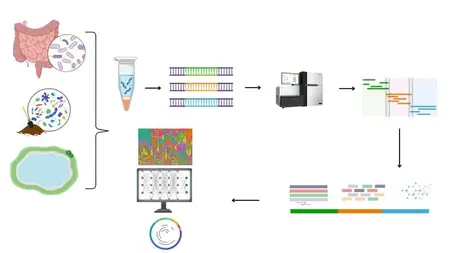
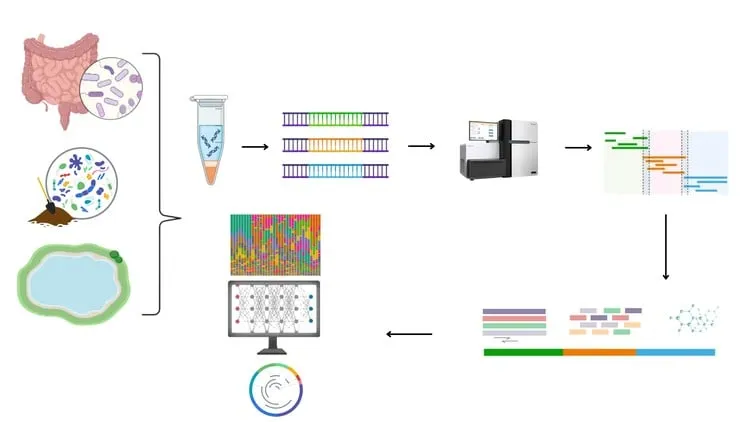
Are you ready to unlock the secrets of microbial communities that live inside us and all around us? Whether you’re a life sciences student, a research scientist, or simply curious about the invisible world of microbes, this course will equip you with the essential skills to perform real-world metagenomics and microbiome data analysis using industry-standard tools likeQIIME2, Kraken2, Bracken, and Linux-based pipelines.
"Master Metagenomics and Microbiome Data Analysis Using Linux"is a comprehensive, hands-on course designed to take you from raw sequencing data to meaningful biological insights. With a practical approach and clear guidance, this course will help you navigate the complex but exciting world of microbial community profiling using both16S rRNA gene sequencingandwhole-genome shotgun metagenomics.
You will start by understanding the fundamentals of metagenomics, microbiomes, and how they are transforming fields like human health, agriculture, and environmental science. You’ll explore the difference between 16S rRNA sequencing and shotgun sequencing, and learn where and how to access public datasets for your own analysis.
Next, you’ll set up yourLinux-based bioinformatics environment, either through WSL (Windows Subsystem for Linux) or VirtualBox, and get comfortable using basic Linux commands that are crucial for data preprocessing, file management, and running bioinformatics tools.
From there, we dive deep intoQIIME2, one of the most widely used platforms for microbiome analysis. You'll learn how to import your sequencing data, perform quality control, denoise reads using DADA2, and generate interactive visualizations. We’ll walk you through computingalpha and beta diversity,taxonomic classification,differential abundance testing, and how to create publication-quality plots that explain microbial community structure and variation.
But we don’t stop at 16S. This course also takes you into the powerful world ofshotgun metagenomicsusingKraken2, Bracken, andKronafor high-resolution taxonomic classification and microbial abundance estimation. You'll learn how to download and configure metagenomic databases, classify reads, and visualize your results in interactive formats. These tools are crucial for researchers working in microbiome profiling, diagnostics, drug discovery, and microbial ecology.
Throughout the course, you'll get access to:
Pre-formatted sample datasets
Scripts and command templates
Metadata files and manifest examples
Hands-on walkthroughs of every step
Visualization tools and interpretation strategies
This course is built forbeginners and intermediate learners. You don’t need prior experience in bioinformatics or command-line tools—just a willingness to learn and explore.
By the end of the course, you'll be able to confidently:1. Set up a working bioinformatics environment2. Analyze 16S and shotgun metagenomic datasets3. Interpret taxonomic and functional profiles4. Perform diversity and differential abundance analysis5. Generate high-quality reports and plots
If you're planning a research project, working in a lab, or aiming to publish microbiome data, this course gives you the technical skills and practical confidence you need.
Join nowand become part of a growing community exploring the power of microbiome data in health, environment, and beyond!
Who this course is for:
- Life sciences and biology students who want to get started in bioinformatics or microbiome research
- Lab technicians and clinical researchers interested in microbial profiling and diagnostics
- Academic researchers and graduate students working on microbiome or metagenomics projects
- Bioinformatics enthusiasts and self-learners eager to understand microbial communities using sequencing data
- Healthcare and environmental science professionals exploring the role of microbiomes in health, agriculture, and ecology
- Anyone with a curious mind who wants to learn QIIME2, Kraken2, and Linux pipelines to analyze microbiome datasets
More Info